ABSTRACT
Automatically acquired and reconstructed 3D breast ultrasound images allow radiologists to detect and evaluate breast lesions in 3D. However, assessing potential cancers in 3D ultrasound can be difficult and time consuming. In this study, we evaluate a 3D lesion segmentation method, which we had previously developed for breast CT, and investigate its robustness on lesions on 3D breast ultrasound images. Our dataset includes 98 3D breast ultrasound images obtained on an ABUS system from 55 patients containing 64 cancers.
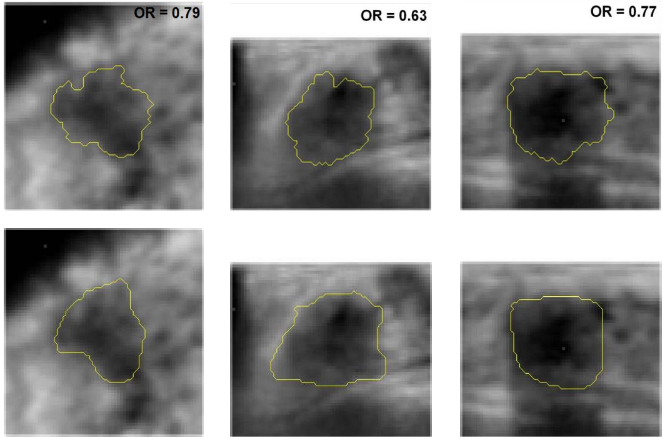
Cancers depicted on 54 US images had been clinically interpreted as negative on screening mammography and 44 had been clinically visible on mammography. All were from women with breast density BI-RADS 3 or 4. Tumor centers and margins were indicated and outlined by radiologists. Initial RGI-eroded contours were automatically calculated and served as input to the active contour segmentation algorithm yielding the final lesion contour. Tumor segmentation was evaluated by determining the overlap ratio (OR) between computer-determined and manually-drawn outlines. Resulting average overlap ratios on coronal, transverse, and sagittal views were 0.60 ± 0.17, 0.57 ± 0.18, and 0.58 ± 0.17, respectively.
All OR values were significantly higher the 0.4, which is deemed “acceptable”. Within the groups of mammogram-negative and mammogram-positive cancers, the overlap ratios were 0.63 ± 0.17 and 0.56 ± 0.16, respectively, on the coronal views; with similar results on the other views. The segmentation performance was not found to be correlated to tumor size. Results indicate robustness of the 3D lesion segmentation technique in multi-modality 3D breast imaging.
Keywords
Computer-aided diagnosis, 3D segmentation, breast imaging, ultrasound
INTRODUCTION
recently has been indicated for use in breast cancer screening as an adjunct to screening mammography for women with dense breasts [3,4]. Mammographically occult lesions are often small in size and not associated with calcifications. If presenting in dense breasts, it is even more difficult to be visualized on mammography by the radiologists [5,6]. US, however, has been reported for its ability to depict these mammographically occult cancers [7] and increase detection of small cancers as an addition of screening [4,8].
3D coronal reconstructed breast ultrasound images provide not only transverse or sagittal views as conventional ultrasound images but also can display the coronal views of the breasts at different depths. However, interpreting 3D US images may be challenging for some depending on the image noise level and distortion resulting from having different spatial resolution in three dimensions. Therefore, we expect CAD (computer-aided detection/diagnosis) to help alleviate the burden. In this study, we aim to test the robustness of a 3D lesion segmentation algorithm that was previously developed for breast CT [9]. In addition, a comparison of segmentation results was conducted between two groups of cases -- mammogram-positive and mammogram-negative -- in an additional robustness analysis of the proposed 3D lesion segmentation procedure.
REFERENCES
- [1] Kolb, T. M., Lichy, J. and Newhouse, J. H., “Comparison of the performance of screening mammography, physical examination, and breast US and evaluation of factors that influence them: an analysis of 27,825 patient evaluations,” Radiology 225, 167-175 (2002).
- [2] Leconte, I., Feger, C., Galant, C., Berliere, M., Vande Berg, B., D’Hoore, W. and Maldague, B., “Mammography and subsequent whole-breast sonography of nonpalpable breast cancers: the importance of radiologic breast density,” AJR 180, 1675-1679 (2003).
- [3] Berg, W. A., Blume, J. D., Cormack, J. B., Mendelson, E. B., Lehrer, D., Böhm-Vélez, M., Pisano, E. D., Jong, R. A., Evans, W. A., Morton, M. J., Mahoney, M. C., Larsen, L. H., Barr, R. G., Farria, D. M., Marques, H. S. and Boparai, K., “Combined screening with ultrasound and mammography vs mammography alone in women at elevated risk of breast cancer,” JAMA 299, 2151-2163 (2008).
- [4] Giger, M. L., Miller, D. P., Brown, J. B., Inciardi, M. F., Metz, C. E., Jiang, Y., Brem, R. F., Nishikawa, R. M., Edwards, A. V. and Papaioannou, J., “Clinical reader study examining the performance of mammography and automatic breast ultrasound in breast cancer screening,” RSNA SSJ01-04 (2012).
- [5] Dershaw D. D., “Breast disease missed by mammography,” Applied Radiology, 26, 24-28 (1997).
- [6] Huynb, P. T., Jarolimek, A. M. and Daye, A., “The false-negative mammogram,” Radiographics 18, 1137-1154 (1998).
- [7] Kolb, T. M., Lichy, J. and Newhouse, J. H., “Occult cancer in women with dense breasts: detection with screening US – diagnosis yield and tumor characteristics,” Radiology 207, 191-199 (1998).
- [8] Kolb, T. M., Lichy, J., and Newhouse, J. H., “Comparison of the performance of screening mammography, physical, examination, and breast US and evaluation of factors that influence them: an analysis of 27,825 patient evaluations,” Radiology 225, 165-175 (2002).
- [9] Kuo, H., Giger, M. L., Reiser, I., Boone, J. M., Lindfors, K. K., Yang, K. and Edwards, A., “Evaluation of stopping criteria for level set segmentation of breast masses in contrast-enhanced dedicated breast CT,” Proc. SPIE 8315, 83152C (2012).
- [10]Reiser, I., Joseph, S. P., Nishikawa, R. M., Giger, M. L., Boone, J. M., Lindfors, K. K., Edwards, A., Packard, N. Moore, R. H., and Kopans, D. B., “Evaluation of a 3D lesion segmentation algorithm on DBT and breast CT images,” Proc. SPIE 7624, 119-129 (2010).
- [11]Kupinski, M. A. and Giger, M. L., “Automated seeded lesion segmentation on digital mammograms,” IEEE Trans. Med. Imaging 17, 510-517 (1998).
- [12]Li, C., Xu, C., Gui, C. and Fox M. D., “Level set evolution without re-initialization: a new variational formulation,” Proc. IEEE CVPR’05 1, 430-436 (2005).
- [13]Yuan, Y., Giger, M. L., Li, H., Suzuki, K. and Sennett, C., “A dual-stage method for lesion segmentation on digital mammograms,” Med. Phys. 34, 4180-4193 (2007).
- [14]Zou, K. H., Warfield, S. K., Bharatha, A., Tempany, C. M. C., Kaus, M. R., Haker, S. J., Wells, W. M., Jolesz, F. A. and Kinkinis, R., “Statistical validation of image segmentation quality based on a spatial overlap index,” Acad. Radiol. 11, 178-189 (2004).
- [15]Chen, W., Giger, M. L. Bick, U., “A fuzzy c-means (FCM)-based approach fo computerized segmentation of breast lesions in dynamic contrast-enhanced MR images,” Acad. Radiol. 13, 63-72 (2006).
Preview Journals